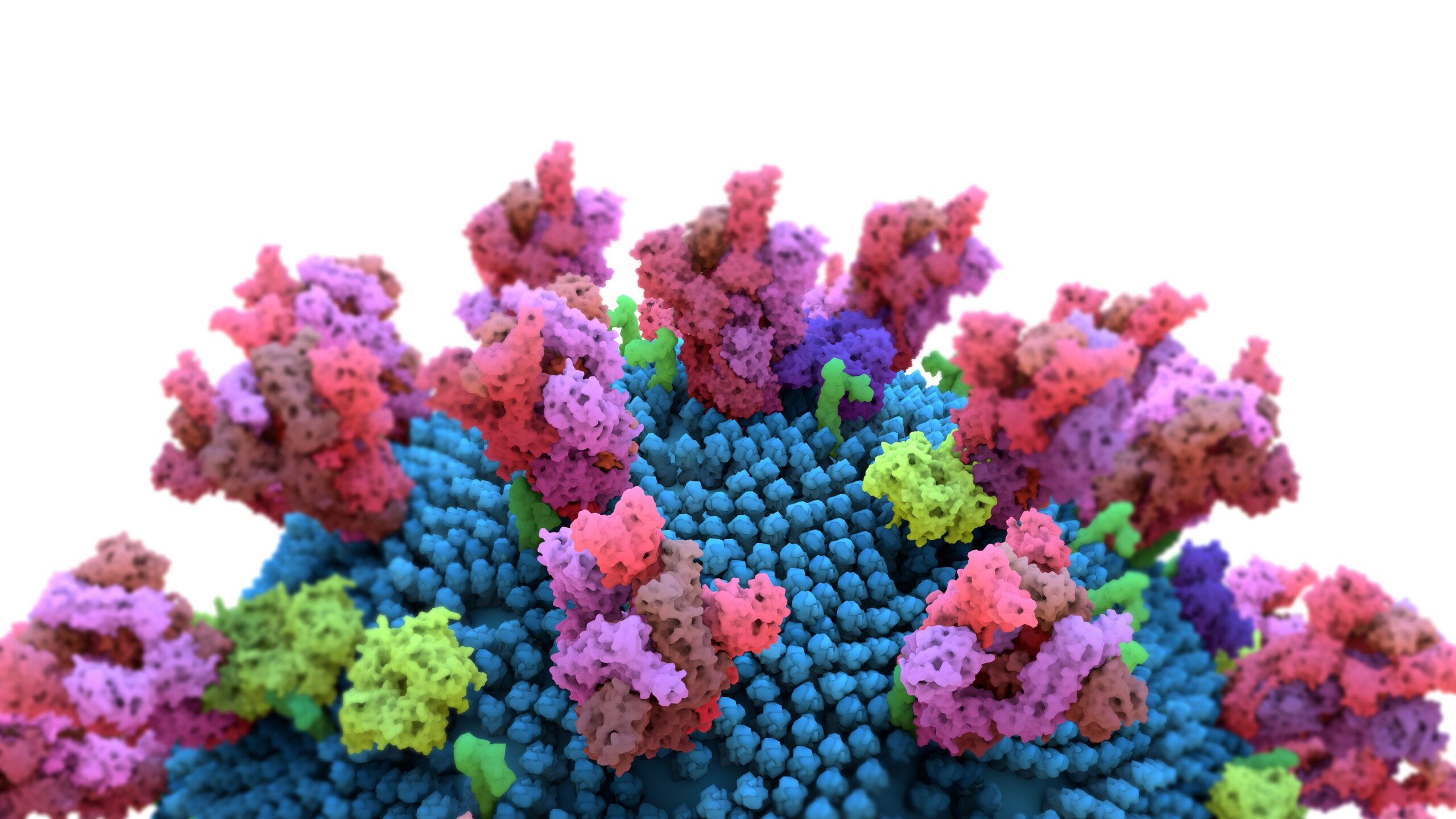
Sugar particles form the protective covering for the virus – detecting attack points – the practice of healing

The Max Planck Institute identifies a new attack point for the Coronavirus
A German research team used a new analysis method to detect a previously unknown attack point in the SARS-CoV-2 coronavirus spike protein. The team showed that some sugar molecules protect the spike protein and protect a large portion of the immune defense cells. However, the team also discovered less protected areas of spike protein that could be targeted.
Researchers at the Max Planck Institute for Biophysics in Frankfurt am Main were able to use a dynamic model of SARS-CoV-2 to understand the protective function of the virus, which protects spike proteins from immune defense cells. At the same time, the investigation also revealed weaknesses in the virus shield. The results were recently published in the popular magazine.Plus” Foot.
Spike protein as a major feature of SARS-CoV-2
As the working group emphasizes, the spike protein is a key feature of the SARS-CoV-2 coronavirus, as it can cling to cell surfaces with the help of the protein in order to subsequently infect them. Extensive research has made it possible to create detailed models of the Corona virus and its prickly protein. Researchers at the Max Planck Institute have now refined these models.
Previous embodiments of the spike protein were stable and could not represent movement. For the first time, the new model is able to simulate the motions of the spiky protein itself and the glycan chains around it.
Like a windshield wiper
The simulations show that the sugar particles on the spike protein act as a dynamic protective shield that helps the virus evade the human immune system. The researchers compared the protection function with the windshield wiper that cleans a car’s windshield. The glycans move back and forth on the spike protein, preventing neutralizing antibodies from attaching to the spike protein.
Not all places are equally protected
However, the research also showed that not all sites are well protected. Similar to a windshield wiper, the sugar particles do not cover all areas of the spike protein. Researchers confirmed that some areas are less protected by the glycan shield than others. Some areas detected were already identified as weaknesses in previous studies, and others were not yet known.
New starting points against SARS-CoV-2 mutations
“We are in a phase of an epidemic that is constantly changing due to the emergence of new variants of SARS-CoV-2, with mutations specifically focused in the protein spike,” explains Matthews Secura of the research team. The new approach could support the design of vaccines and therapeutic antibodies, especially if approaches actually fail.
Discovering weaknesses in viral proteins
At the same time, the method developed also represents a new way to find potential vulnerabilities on other viral proteins, and this sums up the research team at the Max Planck Institute. Only recently, an American research team discovered another vulnerability in the coronavirus spike protein: You can learn more about this in the article:COVID-19: Identifying a new virus vulnerability(Fb)
Author and source information
This text complies with the requirements of the specialized medical literature, medical guidelines and current studies and has been examined by medical professionals.
author:
Diploma Editor (FH) Volker Plasik
Inflated:
- Max Planck Society: The dynamic model of the spiky SARS-CoV-2 protein shows targets for new vaccines (Posted: 04/01/2021), mpg.de
- Matthews Secura, Suren von Bülow, Florian EC Planck, et al: Computational Loop Map of the Spike SARS-CoV-2 in: Plos Computational Biology, 2021, Journal.plos.org
important note:
This article is for general guidance only and is not intended to be used for self-diagnosis or self-medication. He cannot replace a visit to the doctor.

“Organizer. Social media geek. General communicator. Bacon scholar. Proud pop culture trailblazer.”
